General protocol for PCR on difficult templates

- Reaction setup
A typical KAPA2G Robust reaction consists of the following:

- 1X GC Buffer + 4%DMSO.
- 1X Buffer A + 5%DMSO + 1X KAPA Enhancer 1.
- Cycling parameters
Notes on reaction setup:
1. Reaction volumes of 10 - 50μl are recommended. For volumes larger or smaller than 25μl, scale reagents listed in the table up or down proportionally.
2. Ensure that all components arefully thawed before use. Vortex KAPA2G Buffers and KAPA Enhancer 1 before each use.
3. All three 5X KAPA2G Buffers contain MgCl2. Use buffers at a final concentration of 1X (1.5 mM MgCl2). If a particular assay requires more MgCl2, supplement the reaction with the MgCl2 supplied in the kit. The optimal MgCl2 concentration for each application should be determinedempirically in a MgCl2 gradient PCR.
4. KAPA2G Buffer A is the recommended buffer for templates or amplicons with a GC content <65%. It has been optimized for the KAPA2G Robust enzyme and offers high yields, specificity and sensitivity.
5. Buffer B has a very different composition to Buffer A and may work better for some amplicons, particularly when samples are contaminated with anionic inhibitors. It is the recommended buffer for Colony PCR. For problematic assays, first evaluate both Buffer A and Buffer B before attempting further optimization.
6. KAPA2G GC Buffer is specifically formulated for templates or amplicons with a high GC content, or templates that are difficult to amplify as a result of stable secondary structure. For such samples, first try the GC Buffer at 1x concentration without any other additives. For particularly recalcitrant templates/amplicons, try the following:
7. KAPA Enhancer 1 is a proprietary additive that improves
reaction efficiency and specificity for some, but not all
primer-template combinations. It is supplied as a 5X solution and
should always be used at a final concentration of 1X. For problematic
assays, first try Buffer A or Buffer B, with or without 1X KAPA
Enhancer 1, before attempting further optimization. The GC Buffer may
also be tried for problematic assays, even if the GC content of the
template or amplicon is <65%. Do not combine KAPA Enhancer 1 with
the GC Buffer.
8. 0.5 units KAPA2G Robust DNA Polymerase per 25μl reaction should be sufficient for most assays. For GC rich templates, double the amount of enzyme (1 unit per 25μl reaction) is likely to improve results. The amount of enzyme may also be increased for crude samples, samples containing inhibitors and the amplification of longer amplicons. If smearing or a high background of non-specific amplicons occurs, reduce the amount of enzyme.
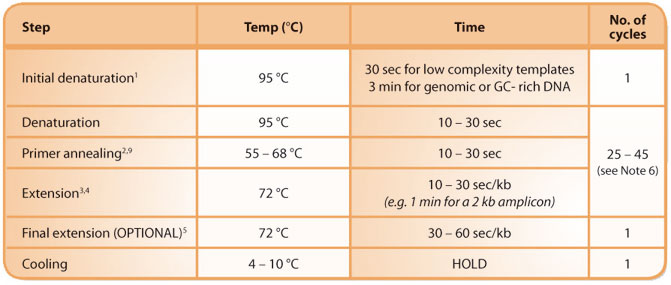
A typical KAPA2G Robust reaction consists of the following:
- >106 copies 25 cycles
- >104 - 106 copies 30 cycles
- <104 copies 35 cycles
Notes on cycling parameters:
1. For recalcitrant templates, the initial denaturation may be increased to a maximum of 10 min.
2. For primers with an optimal annealing temperature (Ta) between 68 and 72 °C, a 2-step protocol with a combined annealing/ extension step of 45 - 75 sec/kb at 68– 72 °C may be used.
3. 30 sec/kb extension time per cycle should be sufficient for most applications. For difficult templates or samples, this may be extended to 1 min/kb.
4. For AT rich templates and amplicons, extension may be performed at 68 °C.
5. A final extension is only necessary if PCR products are to be cloned into TA-cloning vectors.
6. The number of cycles depends on the amount of starting material (target copy number) in the reaction. The following may be used as a general guideline:

- Reduce the annealing time to a maximum of 15 sec per cycle.
- Reduce the extension time to 15 sec/kb.
- Reduce the number of cycles.
- Optimize the Ta for the specific template-primer combination in a Ta gradient PCR.
7. If a very high yield of the target amplicon is obtained or if smearing or non-specific amplification occurs, try one or more of the following:
8. For amplification from crude samples, e.g. Colony PCR, use 5 min initial denaturation (95 °C) and 30 sec denaturation per cycle. 15 sec annealing per cycle should be sufficient in most cases. The optimal extension rate will depend on the nature of the sample and assay.
9. When designing primers, the theoretical melting temperature (Tm) of primers used together should be matched as closely as possible. As a first approach, use an annealing temperature (Ta) 3– 5 °C lower than the lowest Tm of the two primers. For best performance, the optimal Ta for a primer pair should be determined empirically by Ta gradient PCR. Because primer melting characteristics are affected by the chemical environment, the optimal Ta for a specific primer pair should be determined in the PCR buffer used for the assay and may differ from one buffer system to another. Sample composition may also affect primer annealing, particularly if high levels of inhibitors are present. For optimal results with KAPA2G Robust, primers with annealing temperatures <55 °C are not recommended.

- Reaction setup and cycling parameters
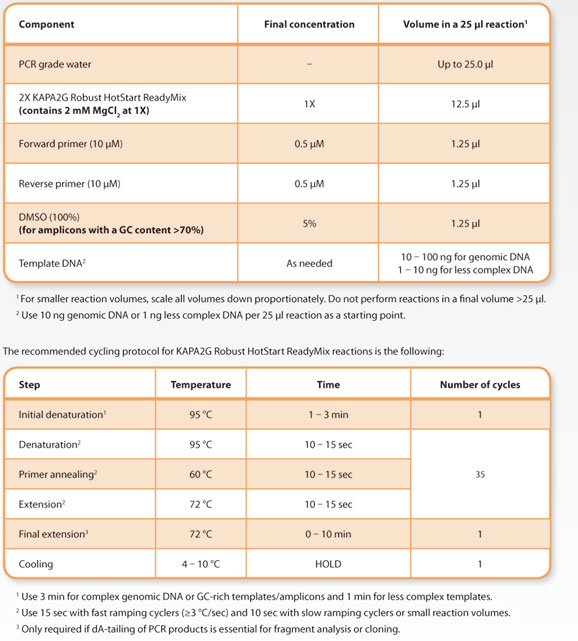
The standardized reaction setup recommended for KAPA2G Robust HotStart ReadyMix reactions is the following:
- Important parameters
- Cycling parameters
The cycling protocol given in Section 3 has been validated across a wide range of amplicons≤1 kb, with a GC content ranging from 25– 85%. Although the KAPA2G Robust HotStart DNA Polymerase only needs 30 sec at 95 °C for reactivation, it is important to use sufficient denaturation times, particularly for GC-rich templates/amplicons. Annealing time should not be increased or decreased beyond the range of 10– 15 sec/cycle, as this may promote non-specific amplification and smearing or low reaction efficiency. To improve yields, rather increase the extension time, template concentration and/or number of cycles. An annealing temperature of 60 °C is recommended as the starting point for routine PCR using a single, standardized protocol. However, depending on the characteristics of a particular set of amplicons to be processed together, this may be adjusted in the range of 55 °C– 65 °C. Annealing temperatures <55 °C are not recommended. To improve yields of problematic amplicons, the extension time may be increased to 30 sec per cycle. For subsets of“easy” amplicons, the extension time may be reduced as low as 5 sec/cycle. - Amplification of GC-rich and other problematic amplicons
It is recommended that reactions are supplemented with DMSO, to a final concentration of 5%, for the amplification of fragments with a GC content >70%. Any PCR-grade DMSO solution may be used. For particularly recalcitrant amplicons, a final DMSO concentration of up to 7.5%may be used. KAPA2G Robust HotStart ReadyMix may also be used in conjunction with 1X KAPA Enhancer 1, a proprietary PCR additive supplied in KAPA2G Robust and KAPA2G Robust HotStart PCR Kits. This has been shown to improve yields and specificity with GC-rich and other problematic amplicons. If optimization of the template concentration, MgCl2 concentration, DMSO concentration and/or annealing temperature does not yield satisfactory results with a particular primer-template combination, a KAPA2G Robust HotStart PCR Kit is recommended for further optimization. In these kits, KAPA2G Robust HotStart DNA Polymerase is supplied separately, with three proprietary KAPA2G reaction buffers and KAPA Enhancer 1, offering a wide range of optimization - Primers
Primer design and quality can have a significant effect on the success rates achieved in routine PCR employing a single, standardized reaction setup and cycling protocol. Primers should be carefully designed to eliminate the possibility of primerdimer formation and spurious annealing as far as possible, and should be suitable for use with a single annealing temperature in the range of 55– 60 °C. Always dilute and store primers in a buffered solution (e.g. 10 mM Tris-HCl, pH 8.0– 8.5) instead of PCR-grade water. Good primer quality is particularly important for consistent and successful amplification of GC-rich fragments. - 4. 5 MgCl2 concentration
KAPA2G Robust HotStart ReadyMix contains MgCl2 at a 1X concentration of 2 mM, which has been determined to be optimal for the amplification of diverse amplicons using a standardized protocol. If more MgCl2 is required for a specific primertemplate combination or assay, reactions may be supplemented with any PCR-grade MgCl2 solution. Add 0.5μl of a 25 mM MgCl2 solution to increase the final MgCl2 concentration in a 25μl reaction by 0.5 mM. The optimal MgCl2 concentration for a specific assay may be determined empirically in a MgCl2 gradient PCR. - Troubleshooting
A final concentration of 0.5μM of each primer is recommended for routine PCR using a single, standardized reaction setup.However, primer concentration may be optimized in the range of 0.1– 1μM for specific assays.


