2×TransTaq® High Fidelity (HiFi) PCR SuperMix I (-dye)
Referentie AS131-01
Formaat : 1ml
Merk : TransGen Biotech
Human GM-CSF ELISA Kit
Catalog Number: NE105
Price:Please inquire first
Product Details
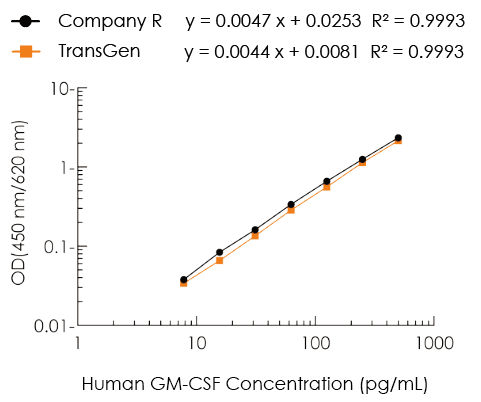
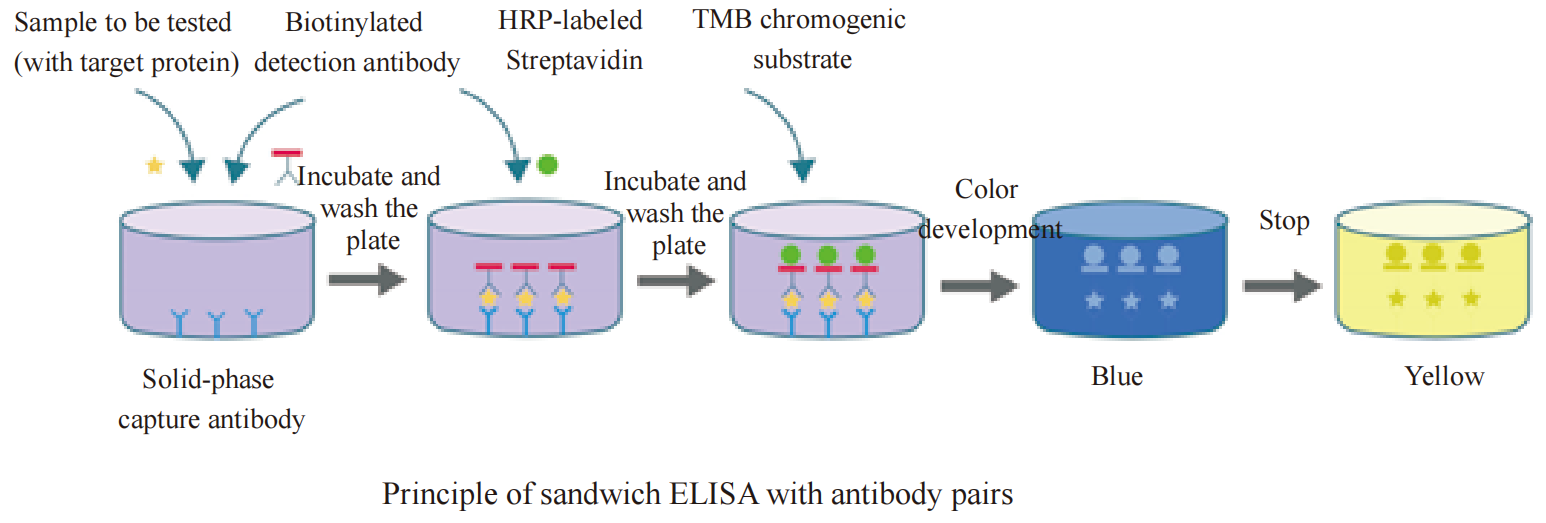
Human granulocyte-macrophage colony-stimulating factor, also known as GM-CSF, or CSF-2, is a monomeric glycoprotein secreted by a variety of activated immune cells, mesenchymal cells and epithelial cells. The amino acid sequence homology of mature human GM-CSF with mouse and rat GM-CSF is 54% and 63%, respectively. GM-CSF has multiple biological functions, such as the ability to stimulate the differentiation of CD34+ stem cell precursors into monocytes, neutrophils and eosinophils; it can act as a chemokine for neutrophils and dendritic cells. GM-CSF is also involved in the autoimmune response mediated by Th1 and Th17 cells and is related to the inflammatory activation of dendritic cells, microglia, alveolar macrophages and eosinophils. In addition, GM-CSF regulates tumor cell proliferation, invasion and metastasis through immune-dependent and independent pathways. This kit uses the Sandwich Enzyme-Linked Immunosorbent Assay (ELISA) to measure the amount of GM-CSF in human serum, plasma, and cell culture supernatant. The ELISA microplate in the kit is precoated with a high-affinity anti-human GM-CSF antibody. The standard or test sample and a biotin-labeled anti-human GM-CSF detection antibody are added to the microplate wells at the same time. After incubation, GM-CSF present in the sample will be bound specifically to the precoated antibody and the detection antibody. After washing, a horseradish peroxidase-labeled streptavidin (Streptavidin-HRP) is added to the microplate wells and incubated. Biotin and streptavidin on the detection antibody produces the "coated antibody-human GM-CSF protein-detection antibody-Streptavidin-HRP" immune complex through high-strength non-covalent binding. After washing again, the chromogenic substrate TMB is added to the wells. HRP catalyzes the TMB substrate to create blue coloration, with intensity positively correlated with the concentration of GM-CSF in the sample. The reaction is stopped by adding stop solution, and the absorbance is measured at 450 nm (reference wavelength 570-630 nm). A standard curve is drawn to calculate the concentration of GM-CSF in the sample based on the absorbance value. This kit is highly specific, has high detection sensitivity and is convenient to use.
Storage
at 2~8°C in the dark for one year
Shipping
ice bag (4℃)