HSPA4 Antibody
Referentie OASE00031
Formaat : 100ug
Merk : Aviva Systems Biology
| Datasheets/Manuals | Printable datasheet for anti-HSPA4 (OASE00031) antibody |
|---|
| Predicted Species Reactivity | Plant |
|---|---|
| Clonality | Monoclonal |
| Clone | 5G1-95 |
| Isotype | IgG1 |
| Host | Mouse |
| Conjugation | Unconjugated |
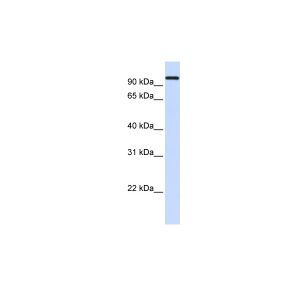
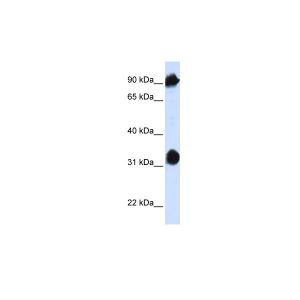
| Application | ELISA, WB |
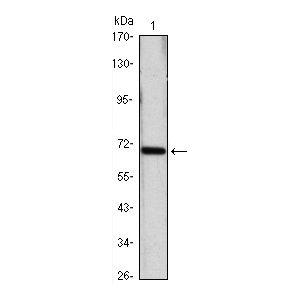
| Additional Information | Background Info: Detects ~70kDa. Recognizes constitutive and inducible plants Hsp70 (Hsc70/Hsp72). |
| :: | Scientific Background: Hsp70 genes encode abundant heat-inducible 70-kDa hsps (hsp70s). In most eukaryotes hsp70 genes exist as part of a multigene family. They are found in most cellular compartments of eukaryotes including nuclei, mitochondria, chloroplasts, the endoplasmic reticulum and the cytosol, as well as in bacteria. The genes show a high degree of conservation, having at least 50% identity (1). The N-terminal two thirds of hsp70s are more conserved than the C-terminal third. Hsp70 binds ATP with high affinity and possesses a weak ATPase activity which can be stimulated by binding to unfolded proteins and synthetic peptides (2). When hsc70 (constitutively expressed) present in mammalian cells was truncated, ATP binding activity was found to reside in an N-terminal fragment of 44 kDa which lacked peptide binding capacity. Polypeptide binding ability therefore resided within the C-terminal half (3). The structure of this ATP binding domain displays multiple features of nucleotide binding proteins (4). All hsp70s, regardless of location, bind proteins, particularly unfolded ones. The molecular chaperones of the hsp70 family recognize and bind to nascent polypeptide chains as well as partially folded intermediates of proteins preventing their aggregation and misfolding. The binding of ATP triggers a critical conformational change leading to the release of the bound substrate protein (5). The universal ability of hsp70s to undergo cycles of binding to and release from hydrophobic stretches of partially unfolded proteins determines their role in a great variety of vital intracellular functions such as protein synthesis, protein folding and oligomerization and protein transport. |
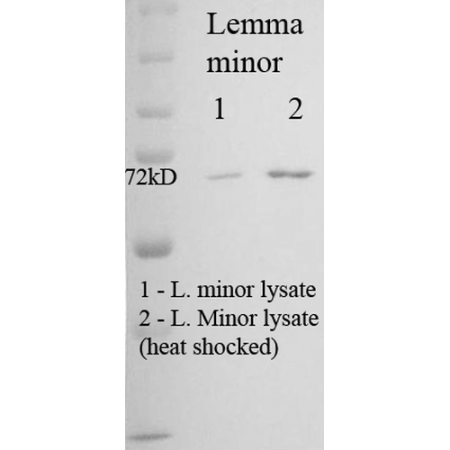
| :: | Certificate of Analysis: 1 ug/mL was sufficient for detection of Hsp70/Hsc70 in 20ug of Phaseolus aureus (mung bean) lysate by colorimetric immunoblot analysis using Goat anti-mouse IgG:HRP as the secondary antibody. |
| Reconstitution and Storage | Store at -20C. Shipping with Blue Ice or 4C. |
| Immunogen | Purified HSP70 from Phaseolus aureus (mung bean) |
| Purification | Protein G Purified |
| Concentration | 1 mg/ml |
| Specificity | Detects ~70kDa. Recognizes constitutive and inducible plants HSP70 (HSC70/HSP72). Does not cross-react with Human, Rat, bacteria (DNAK) or Human Bip. |
| Dilution | WB (1:1000); optimal dilutions for assays should be determined by the user. |
| Storage Buffer | PBS pH7.4, 50% glycerol, 0.09% sodium azide |
| Description | HSP70 genes encode abundant heat-inducible 70-kDa HSPs (HSP70s). In most eukaryotes HSP70 genes exist as part of a multigene family. They are found in most cellular compartments of eukaryotes including nuclei, mitochondria, chloroplasts, the endoplasmic reticulum and the cytosol, as well as in bacteria. The genes show a high degree of conservation, having at least 50% identity (1). The N-terminal two thirds of HSP70s are more conserved than the C-terminal third. HSP70 binds ATP with high affinity and possesses a weak ATPase activity which can be stimulated by binding to unfolded proteins and synthetic peptides (2). When HSC70 (constitutively expressed) present in mammalian cells was truncated, ATP binding activity was found to reside in an N-terminal fragment of 44 kDa which lacked peptide binding capacity. Polypeptide binding ability therefore resided within the C-terminal half (3). The structure of this ATP binding domain displays multiple features of nucleotide binding proteins (4). All HSP70s, regardless of location, bind proteins, particularly unfolded ones. The molecular chaperones of the HSP70 family recognize and bind to nascent polypeptide chains as well as partially folded intermediates of proteins preventing their aggregation and misfolding. The binding of ATP triggers a critical conformational change leading to the release of the bound substrate protein (5). The universal ability of HSP70s to undergo cycles of binding to and release from hydrophobic stretches of partially unfolded proteins determines their role in a great variety of vital intracellular functions such as protein synthesis, protein folding and oligomerization and protein transport. For more information visit our HSP70 Scientific Resource Guide at http://www.HSP70.com. |
| Reference | 1. Boorstein W. R., Ziegelhoffer T. & Craig E. A. (1993) J. Mol. Evol.38 (1): 1-17. 2. Rothman J. (1989) Cell 59: 591-601. 3. DeLuca-Flaherty et al. (1990), Cell 62: 875-887. 4. Bork P., Sander C. & Valencia A. (1992) Proc. Nut1 Acad. Sci. USA 89: 7290-7294. 5. Fink A.L. (1999) Physiol. Rev. 79: 425-449. |
|---|---|
| Gene Symbol | HSP70/HSC70 |
| Gene Full Name | Heat shock 70 kDa protein 4 |
| Alias Symbols | HSC54, HSC70, HSC71, HSP70 1, HSP701/HSP70 2, HSP70.1, HSP71, HSP72, HSP73, HSPA1, HSPA10, HSPA1A, HSPA1B, LAP1, NIP71 |
| NCBI Gene Id | 106769403 |
| Protein Name | Heat shock 70 kDa protein 4 |
| Uniprot ID | A0A1S3UWN7 |
| Protein Accession # | XP_014510496.1 |