Goat anti-Rabbit IgG (H&L), HRP conjugated
Referência AS09602
Tamanho : 1mg
Marca : Agrisera
Goat anti-Rabbit IgG (H&L), HRP conjugated
AS09 602 | Clonality: Polyclonal | Host: Goat | Reactivity: Rabbit IgG (H&L)
- Shelby Hotz | 2023-03-21For my lab's study, we used this antibody for western blots to detect the abundance of Hsp70 in heat shocked and non-heat shocked bull kelp (Nereocystis leutkeana). We used a 1:5000 for the initial dilution and then a 1:2500 dilution after adding glycerol. Both were done in a 5% Nonfat Dried Milk in 1x Tris Buffered Saline solution. The results were positive, with the protein bands showing up clearly and at the correct weight according to the positive control we used.I am happy to report the antibody has been working effectively and as intended in our molecular research. The instructions that come with the product are very comprehensive and relatively easy to follow, and doing so has allowed us to effectively use the materials as expected and needed. We also appreciate the promptness of the delivery and arrival of the materials, and having the antibody in a lyophilized form is very much appreciated.Thank you for your service.Vasundara Sundar | 2022-12-28We are very satisfied with the goat anti-rabbit antibodies, it worked well with all of the primary antibodies we used. 1:10000 dilution worked well for us.Annika Brünje | 2021-01-19This antibody gives very good results in my Western blot analyses and can be also used in combination with primary antibodies that are a bit more tricky in their application. Usually, I use it in a dilution of 1:10000 to detect proteins in Arabidopsis leaf extracts.Soo Yeon Ko | 2020-05-28I was satisfied with goat anti-rabbit IgG antibodies on protein extracts from Oryza Sativa in dilution : 1: 25 000?udmila Holubová | 2019-10-04We are happy with this antibody, works well with all primary antibodies we used. Our dilution was 1:12500.Ella Nukarinen | 2018-05-23We are very happy with goat anti-rabbit IgG HRP antibody. We use it for Western blots in dilution 1:10 000-1:20 000 for detecting endogenous proteins from Arabidopsis protein extracts.Beata Kmiec | 2017-02-13We're very happy with the antibodies. Our regular application is in western blots with Arabidopsis thaliana total protein extract and with isolated mitochondria and chloroplasts, but we've used it with success in WB with yeast and human samples. We use it in dilution 1:20000.Malgorzata Kwasniak-Owczarek | 2016-11-02We are very satisfied with goat anti-rabbit IgG antibodies on protein extracts from Arabidopsis thaliana in dilution: 1: 25 000.Mirka Herbstova | 2016-04-22I used Goat-anti rabbit IgG HRP conjugated secondary antibody (AS 09 602) at a dilution 1:50 000 for alga species Phaeodactylum and Trachydiscus without cross-reactions.Jakub Dolata, Adam Mickiewicz University, Poznan, Poland | 2014-03-12The goat anti-rabbit IgG-HRP from Agrisera are very nice working antibodies with no unspecific signals in different dillutions (1:50000, 1:100000). We are using them in case of plant (Arabidopsis, Solanum) and human cells extracts as well.Jakub DOlata | 2014-03-12The goat anti-rabbit IgG-HRP from Agrisera are very nice working antibodies with no unspecific signals in different dillutions (1:50000Gerhard Obermeyer, Univ Salzburg, Austria | 2011-11-11We were looking for a 2nd anti-rabbit antibody which does not cross-react with proteins from a crude lily pollen lysate. The goat anti-rabbit IgG-HRP from Agrisera was the only one which showed no unspecific signals and could be used in a 1:40,000 dilution. All other antibodies from 5 different sources cross-reacted with a high number of proteins in the pollen lysate.
| Immunogen: | Purified Rabbit IgG, whole molecule, |
| Host: | Goat |
| Clonality: | Polyclonal |
| Purity: | Immunogen affinity purified using solid phase rabbit IgG. |
| Format: | Lyophilized |
| Quantity: | 1 mg |
| Reconstitution: | For reconstitution add 1.1 ml of sterile water. Let it stand 30 minutes at room temperature to dissolve. Spin centrifuge shortly to remove any particles. Prepare fresh working dilutions daily |
| Storage: | Store lyophilized material at 2-8°C. For long time storage after reconstitution, dilute the antibody solution with glycerol to a final concentration of 50% glycerol and store as liquid at -20°C, to prevent loss of enzymatic activity. For example, if you have reconstituted 1 mg of antibody in 1.1 ml of sterile water, add 1.1 ml of glycerol. Such solution will not freeze in -20°C. If you are using a 1:5000 dilution prior to diluting with glycerol, then you would need to use a 1:2500 dilution after adding glycerol. Prepare working dilution prior to use and then discard, Be sure to mix well but without foaming. |
| Tested applications: | ELISA (ELISA), Immunohistochemistry (IHC), Western blot (WB) |
| Recommended dilution: | 1 : 50 000 -1 : 90 000 (ELISA), 1 : 500 -1 : 5000 (IHC), 1: 10 000 -1 : 50 000 (WB) |
| Confirmed reactivity: | Based on IEP, this antibody reacts with: rabbit IgG heavy chains and light chains on all rabbit immunoglobulins |
| Not reactive in: | Non-immunoglobulin rabbit serum proteins |
| 5 µg of total extract from (1) Hordeum vulgaretotal leaf, (2) Zea mays (3) Spinacia oleracea extracted with PEB (AS08 300) were separated on 4-12% NuPage (Invitrogen) LDS-PAGE and blotted 1h to PVDF. Blots were blocked immediately following transfer in 2% ECL Advance blocking reagent (GE Healthcare) in 20 mM Tris, 137 mM sodium chloride pH 7.6 with 0.1% (v/v) Tween-20 (TBS-T) for 1h at room temperature with agitation. Blots were incubated in the primary anti-PsaC antibody (AS04 042) at a dilution of 1: 10 000 for 1h at room temperature with agitation. The antibody solution was decanted and the blot was rinsed briefly twice, then washed once for 15 min and 3 times for 5 min in TBS-T at room temperature with agitation. Blots were incubated in secondary antibody (goat anti-rabbit IgG horse radish peroxidase conjugated, AS09 602, Agrisera) diluted to 1:50 000 in 2% ECL Advance blocking solution for 1h at room temperature with agitation. The blots were washed as above and developed for 5 min with chemiluminescent detection reagent according to the manufacturers instructions. Images of the blots were obtained using a CCD imager (FluorSMax, Bio-Rad) and Quantity One software (Bio-Rad). Exposure time was 30 seconds. Comparison of Agrisera secondary antibody sensitivity 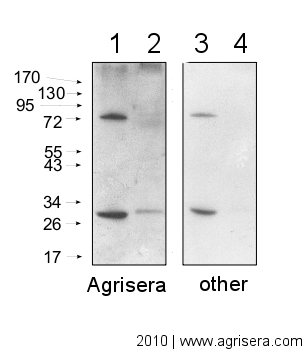 10 μg of mitochondrial fraction from Arabidopsis thaliana (1,3) and Arabidopsis thaliana leaf extract (2,4) were separated on 10% gel and blotted on nitrocellulose membrane using wet transfer (0.22% CAPS, pH 11). Filters where blocked (1.5h) in 5% milk in TBST (1X TBS, 0,1% Tween 20), incubated with 1: 1000 anti-COXII antibodies (2h in TBST) followed by incubation with 1: 10 000 secondary anti-rabbit (1h) HRP-coupled antibodies from Agrisera (left panel) and other manufacture (right panel) and visualized with chemiluminescent detection reagent, on Kodak autoradiography film for 5 s. Antibody in left panel detects target protein also in total cell extract (2) and can be used in higher dilution than applied 1: 10 000. Agrisera goat anti-rabbit HRP conjugated antibody (AS09 602) can be used at following dilutions: 1: 50 000 -1: 90 000 (ELISA), 1 : 75 000 with chemiluminescence detection range of extreme low picogram and 1: 25 000 with chemiluminescence detection reagent of mid femtogram (WB), 1: 500 -1: 5000 (IHC). | |
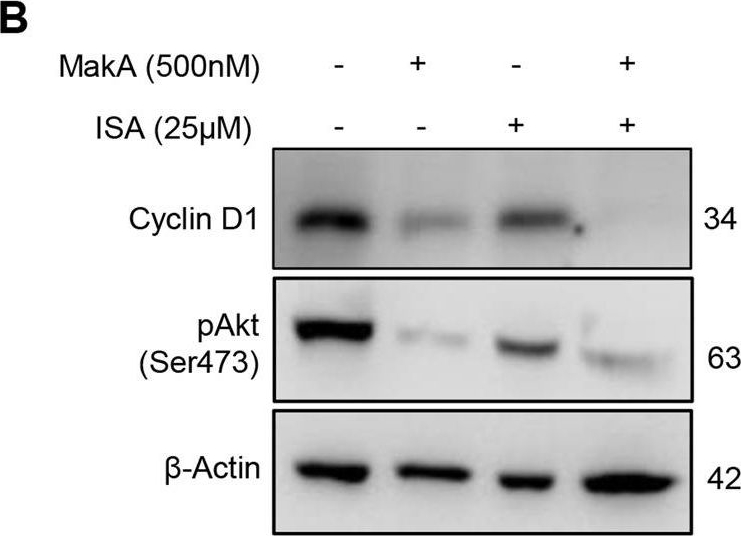
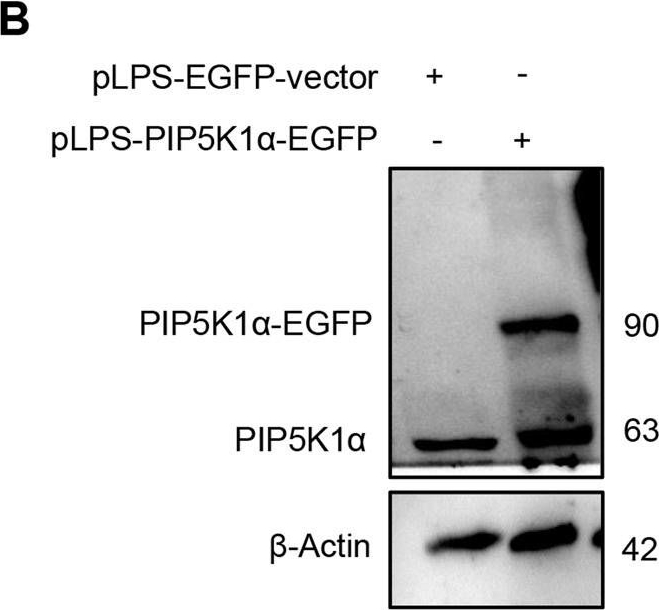
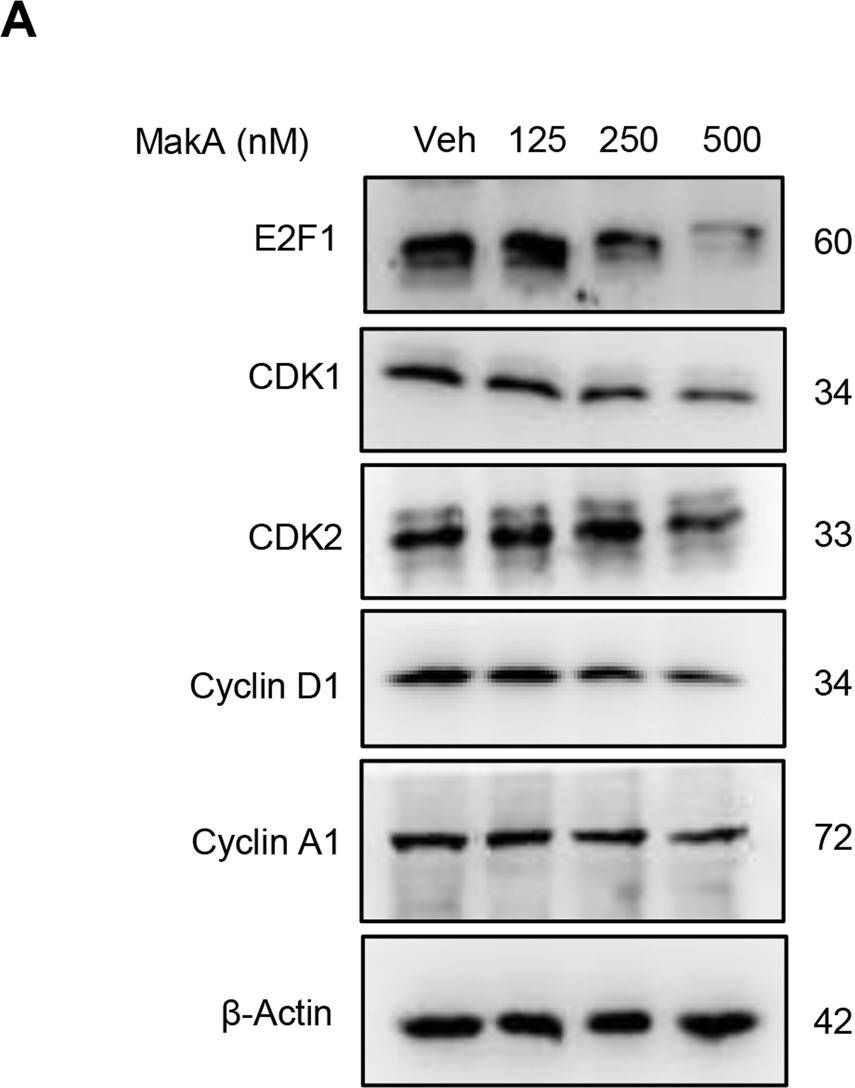
| Application examples: | Reactant: Homo sapiens (Human) Application: Western Blotting Pudmed ID: 36473840 Journal: Cell Death Dis Figure Number: 5B Published Date: 2022-12-06 First Author: Toh, E. Impact Factor: 5.638 Open PublicationMakA inhibits tumour cell proliferation in a PIP5K1α-dependent manner.A MakA and PIP5K1α inhibitor, ISA-2011B synergistically inhibited HCT8 cell proliferation as quantified by colony forming assay. Data are representative of two independent experiments; bar graphs show mean ± s.d. Significance from replicates was determined using a one-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test (post-test) against vehicle (Veh). *p ≤ 0.05 and **p ≤ 0.01. B Western blot analyses of cyclin D1 and pAkt levels in HCT8 cells upon 48 h treatment with MakA (500 nM), ISA-2011B (25 μM) or a combination of MakA (500 nM) and ISA-2011B (25 μM). β-actin was used as a loading control. C Histograms showing the normalized quantification of cyclin D1 and pAkt relative to total actin. Data are representative of at least two independent experiments; bar graphs show mean ± s.d. Significance from replicates was determined using a one-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test (post-test) against vehicle (Veh). *p ≤ 0.05 and **p ≤ 0.01. D Confocal microscopy analysis of vehicle (Veh), MakA (500 nM), ISA-2011B (25 μM), or a combination of MakA (500 nM) and ISA-2011B (25 μM) treated HCT8 cells (48 h treatment). Cells were stained with cell proliferation marker, Ki67 (green). Nuclei were counterstained with DAPI (blue). Inhibition of HCT8 cell proliferation was quantified by an increase in the number of Ki67-negative stained cells from four individual microscopy fields of view (65-165 cells per field of view). *p ≤ 0.05 and **p ≤ 0.01. Scale bar = 20 μm. Reactant: Homo sapiens (Human) Application: Western Blotting Pudmed ID: 36473840 Journal: Cell Death Dis Figure Number: 6B Published Date: 2022-12-06 First Author: Toh, E. Impact Factor: 5.638 Open PublicationEffect of MakA in HCT8 cells with PIP5K1α overexpression and schematic summaries of the MakA effects on PIP5K1α-mediated signalling.A HCT8 cells were transfected with EGFP-vector control (upper panel) or PIP5K1α-EGFP (lower panel) plasmids as described in “Materials and methods”. For co-localization experiments of MakA and PIP5K1α, the transfected HCT8 cells were subsequently treated with Alexa568MakA (250 nM, red) for 48 h. Nuclei were counterstained with DAPI (blue). Graphs on the right show fluorescence intensity profiles of the corresponding images along the dotted white lines. Pearson correlation co-efficient was used for calculation of Alexa568MakA (red) co-localization with EGFP or PIP5K1α-EGFP (green) along the cell membrane. Scale bars = 10 μm. B Western blot analysis of PIP5K1α levels in EGFP-vector control or PIP5K1α-EGFP HCT8 total cell lysates. C Confocal microscopy analysis of vehicle (Veh) or MakA (250 nM) treated (48 h treatment) HCT8 cells transfected with EGFP-vector control or PIP5K1α-EGFP plasmids. Cells were stained with cell proliferation marker, Ki67 (green). Nuclei were counterstained with DAPI (blue). Effects on HCT8 cell proliferation was assessed by quantifying the number of Ki67-negative cells from eleven individual microscopy fields of view (65–200 cells per field of view). Significance was determined using an unpaired t test, ns non-significant. D Schematic representations showing (i) the downstream signalling of PIP5K1α during tumour cell proliferation and survival, (ii) the effect of MakA and ISA-2011B inhibition. Reactant: Homo sapiens (Human) Application: Western Blotting Pudmed ID: 36473840 Journal: Cell Death Dis Figure Number: 4A Published Date: 2022-12-06 First Author: Toh, E. Impact Factor: 5.638 Open PublicationMakA induces G2/M cell cycle arrest in HCT8 cells and alters the expression of cell cycle regulatory proteins.A Western blot analysis shows levels of E2F1, CDK1, CDK2, cyclin D1, and cyclin A in HCT8 cells treated with increasing concentrations of MakA (24 h). B, C Histograms indicate the normalized quantification of E2F1, CDK1, CDK2, cyclin D1, and cyclin A1 relative to total actin. Data is representative of at least two independent experiments and is expressed compared to cells treated with vehicle (20 mM Tris-HCl); bar graphs show mean ± s.d. Significance was determined using a one-way analysis of variance (ANOVA) followed by Dunnett’s post-test in comparison to the vehicle control *p ≤ 0.05 and **p ≤ 0.01. D Cell cycle distribution flow cytometric analysis of HCT8 cells treated with MakA protein (500 nM, 24 h). E Histogram showing the distribution of HCT8 cells during each phase of the cell cycle after treatment with vehicle or MakA. Data are representative of three independent experiments. *p ≤ 0.05 and **p ≤ 0.01. |
| Additional information: | Concentration: 1.0 mg/ml. Antibody is provided in: 10 mM Sodium Phosphate, 0.15 M Sodium Chloride, pH 7.2, 1% BSA (w/v), Protease IgG free, 0.1% (v/v) ProClin 150. Affinity purified antibody is >95% pure, according to SDS-PAGE. This antibody can used on a very wide range of samples from various species including many model plants, algae, diatoms and bacteria. |
| Background: | Goat anti-rabbit IgG (H&L) is a secondary antibody conjugated to HRP which binds to all rabbit IgGs in immunological assays. |
| Selected references: | Boussardon et al. (2025). The atypical proteome of mitochondria from mature pollen grains. Curr Biol . 2025 Jan 21:S0960-9822(24)01705-6. doi: 10.1016/j.cub.2024.12.037. Caballero et al. (2024). Connecting high-resolution 3D chromatin maps with cell division and cell differentiation at the root apical meristem. Plant Cell Rep. 2024 Sep 16;43(10):232. doi: 10.1007/s00299-024-03322-8. Truong et al. (2024). Apo-siderophores promote growth of iron-deficient Arabidopsis plants by mobilizing iron from roots to shoots and reducing oxidative stress in roots. Plant Stress, Volume 12, June 2024, 100488. Martín-Merchán et al. (2024). Arabidopsis AGO1 N-terminal extension acts as an essential hub for PRMT5 interaction and post-translational modifications. Nucleic Acids Res . 2024 May 20:gkae387.doi: 10.1093/nar/gkae387. Miloro et al. (2024). Barley AGO4 proteins show overlapping functionality with distinct small RNA-binding properties in heterologous complementation. Plant Cell Rep. 2024 Mar 13;43(4):96. doi: 10.1007/s00299-024-03177-z. Liu et al. (2023). RBPome identification in egg-cell like callus of Arabidopsis. Biol Chem. 2023 Sep 29;404(11-12):1137-1149.doi: 10.1515/hsz-2023-0195. Chung et al. (2023). An RNA thermometer in the chloroplast genome of Chlamydomonas facilitates temperature-controlled gene expression. Nucleic Acids Res. 2023 Nov 10;51(20):11386-11400. doi: 10.1093/nar/gkad816. Shi et al. (2023). Protocol to identify protein-protein interaction networks in Solanum tuberosum using transient TurboID-based proximity labeling. STAR Protoc. 2023 Sep 20;4(4):102577.doi: 10.1016/j.xpro.2023.102577. Lim et al (2022). Arabidopsis guard cell chloroplasts import cytosolic ATP for starch turnover and stomatal opening. Nat Commun. 2022 Feb 3;13(1):652. doi: 10.1038/s41467-022-28263-2. PMID: 35115512; PMCID: PMC8814037. Miklankova et al. (2022) HYPK promotes the activity of the Nalfa-acetyltransferase A complex to determine proteostasis of nonAc-X2/N-degron-containing proteins. Sci Adv. 2022 Jun 17;8(24):eabn6153. doi: 10.1126/sciadv.abn6153. Epub 2022 Jun 15. PMID: 35704578; PMCID: PMC9200280. Hofmann, Wienkoop & Luthje (2022) Hypoxia-Induced Aquaporins and Regulation of Redox Homeostasis by a Trans-Plasma Membrane Electron Transport System in Maize Roots. Antioxidants (Basel). 2022 Apr 25;11(5):836. doi: 10.3390/antiox11050836. PMID: 35624700; PMCID: PMC9137787. Bychkov et al. (2022) The role of PAP4/FSD3 and PAP9/FSD2 in heat stress responses of chloroplast genes. Plant Sci. 2022 Sep;322:111359. doi: 10.1016/j.plantsci.2022.111359. Epub 2022 Jun 20. PMID: 35738478. Vitale et al. (2021) Light Spectral Composition Influences Structural and Eco-Physiological Traits of Solanum lycopersicum L. cv. 'Microtom' in Response to High-LET Ionizing Radiation. Plants (Basel). 2021 Aug 23;10(8):1752. doi: 10.3390/plants10081752. PMID: 34451797; PMCID: PMC8399554. Larsson et al. (2023) Dynamin2 functions as an accessory protein to reduce the rate of caveola internalization Zhang et al. (2023). Structural insights into a unique PSI-LHCI-LHCII-Lhcb9 supercomplex from moss Physcomitrium patens Tükenmez et al (2023). A Highly Substituted Ring-Fused 2-Pyridone Compound Targeting PrfA and the Efflux Regulator BrtA in Listeria monocytogenes. mBio. 2023 Jun 27;14(3):e0044923. doi: 10.1128/mbio.00449-23 Aguiló-Nicolau et al. (2023) Singular adaptations in the carbon assimilation mechanism of the polyextremophile cyanobacterium Chroococcidiopsis thermalis. Photosynth Res. 2023 May;156(2):231-245. doi: 10.1007/s11120-023-01008-y. Žemaitis et al. (2023 The stem cell-supporting small molecule UM171 triggers Cul3-KBTBD4-mediated degradation of ELM2 domain-harboring proteins. J Biol Chem. 2023 May;299(5):104662. doi: 10.1016/j.jbc.2023.104662. Cembrowska-Lech, Rybak (2023) Nanopriming of Barley Seeds-A Shotgun Approach to Improve Germination under Salt Stress Conditions by Regulating of Reactive Oxygen Species. Plants (Basel). 2023;12(2):405. Published 2023 Jan 15. doi:10.3390/plants12020405 Miernicka et al. (2022) The Adjustment Strategy of Venus Flytrap Photosynthetic Apparatus to UV-A Radiation. Cells. 2022;11(19):3030. Published 2022 Sep 27. doi:10.3390/cells11193033 Boussardon, Bag, Juvany, et al. (2022) The RPN12a proteasome subunit is essential for the multiple hormonal homeostasis controlling the progression of leaf senescence. Commun Biol. 2022;5(1):1043. Published 2022 Sep 30. doi:10.1038/s42003-022-03998-4 Linster et al. (2022). Cotranslational N-degron masking by acetylation promotes proteome stability in plants. Nat Commun. 2022 Feb 10;13(1):810. doi: 10.1038/s41467-022-28414-5. PMID: 35145090; PMCID: PMC8831508. Baena et al. (2022) SNARE SYP132 mediates divergent traffic of plasma membrane H+-ATPase AHA1 and antimicrobial PR1 during bacterial pathogenesis. Plant Physiol. 2022 Jun 27;189(3):1639-1661. doi: 10.1093/plphys/kiac149. PMID: 35348763; PMCID: PMC9237740. Franziska et al. (2022) Auxin application to maize plants at flowering increases abundance and activity of plasma membrane H+-ATPase in developing maize kernels, Journal of Plant Nutrition and Soil Science, Volume 185, Issue 5, October 2022. Ye, Zhou, Zhu, et al. (2022) Inhibition of shoot-expressed NRT1.1 improves reutilization of apoplastic iron under iron-deficient conditions. Plant J. 2022;112(2):549-564. doi:10.1111/tpj.15967 Perera-Castro et al (2022). Limitations to photosynthesis in bryophytes: certainties and uncertainties regarding methodology. J Exp Bot. 2022;73(13):4592-4604. doi:10.1093/jxb/erac191 Lim et al (2022). Arabidopsis guard cell chloroplasts import cytosolic ATP for starch turnover and stomatal opening. Nat Commun. 2022 Feb 3;13(1):652. doi: 10.1038/s41467-022-28263-2. PMID: 35115512; PMCID: PMC8814037. Miklankova et al. (2022) HYPK promotes the activity of the Nalfa-acetyltransferase A complex to determine proteostasis of nonAc-X2/N-degron-containing proteins. Sci Adv. 2022 Jun 17;8(24):eabn6153. doi: 10.1126/sciadv.abn6153. Epub 2022 Jun 15. PMID: 35704578; PMCID: PMC9200280. Hofmann, Wienkoop & Luthje (2022) Hypoxia-Induced Aquaporins and Regulation of Redox Homeostasis by a Trans-Plasma Membrane Electron Transport System in Maize Roots. Antioxidants (Basel). 2022 Apr 25;11(5):836. doi: 10.3390/antiox11050836. PMID: 35624700; PMCID: PMC9137787. Bychkov et al. (2022) The role of PAP4/FSD3 and PAP9/FSD2 in heat stress responses of chloroplast genes. Plant Sci. 2022 Sep;322:111359. doi: 10.1016/j.plantsci.2022.111359. Epub 2022 Jun 20. PMID: 35738478. Vitale et al. (2021) Light Spectral Composition Influences Structural and Eco-Physiological Traits of Solanum lycopersicum L. cv. 'Microtom' in Response to High-LET Ionizing Radiation. Plants (Basel). 2021 Aug 23;10(8):1752. doi: 10.3390/plants10081752. PMID: 34451797; PMCID: PMC8399554. Gucum et al. (2021) A patient-based medaka alg2 mutant as a model for hypo-N-glycosylation. Development. 2021 Jun 1;148(11):dev199385. doi: 10.1242/dev.199385. Epub 2021 Jun 9. PMID: 34106226; PMCID: PMC8217707. Wu et al. (2021). Formation of light-harvesting complex (LHC) II aggregates from LHCII-PSI-LHCI complexes in rice plants under high light. J Exp Bot. 2021 May 3:erab188. doi: 10.1093/jxb/erab188. Epub ahead of print. PMID: 33939808. Wojciechowska et al. (2021) Localization and Dynamics of the Methionine Sulfoxide Reductases MsrB1 and MsrB2 in Beech Seeds. Int J Mol Sci. 2021 Jan 2;22(1):402. doi: 10.3390/ijms22010402. PMID: 33401671; PMCID: PMC7795007. Long et al. (2020). FOXO3 is targeted by miR-223-3p and promotes osteogenic differentiation of bone marrow mesenchymal stem cells by enhancing autophagy. Hum Cell. 2020 Sep 13.doi: 10.1007/s13577-020-00421-y. Khajuria et al. (2020). Photochemical Efficiency Is Negatively Correlated With the Delta 9- Tetrahydrocannabinol Content in Cannabis Sativa L. Plant Physiol Biochem. 2020 Apr 8;151:589-600. doi: 10.1016/j.plaphy.2020.04.003. Ignatov et al. (2020). An mRNA-mRNA Interaction Couples Expression of a Virulence Factor and Its Chaperone in Listeria Monocytogenes. Cell Rep, 30 (12), 4027-4040.e7 Hu et al. (2019). Photoprotection capacity of microalgae improved by regulating the antenna size of light-harvesting complexes. J Appl Phycol (2019). doi.org/10.1007/s10811-019-01969-5 Schober et al. (2019). Organelle Studies and Proteome Analyses on Mitochondria and Plastids Fractions from the Diatom Thalassiosira pseudonana. Plant Cell Physiol. 2019 Jun 10. pii: pcz097. doi: 10.1093/pcp/pcz097. Rethore et al. (2019). Arabidopsis seedlings display a remarkable resilience under severe mineral starvation using their metabolic plasticity to remain self-sufficient for weeks. Plant J. 2019 Mar 22. doi: 10.1111/tpj.14325. Contreras et al. (2019). UV-B shock induces photoprotective flavonoids but not antioxidant activity in Antarctic Colobanthus quitensis (Kunth) Bartl. Environmental and Experimental Botany, Volume 159, March 2019, Pages 179-190. Nikkanen et al. (2018). Multilevel regulation of non-photochemical quenching and state transitions by chloroplast NADPH-dependent thioredoxin reductase. Physiol Plant. 2018 Dec 22. doi: 10.1111/ppl.12914. Migocka et al. (2018). Cucumber metal tolerance protein 7 (CsMTP7) is involved in the accumulation of Fe in mitochondria under Fe excess. Plant J. 2018 Jun 22. doi: 10.1111/tpj.14006. Tong et al. (2018). Delivery of siRNA in vitro and in vivo using PEI-capped porous silicon nanoparticles to silence MRP1 and inhibit proliferation in glioblastoma. J Nanobiotechnology. 2018 Apr 13;16(1):38. doi: 10.1186/s12951-018-0365-y. Nikkanen et al. (2018). Regulation of chloroplast NADH dehydrogenase-like complex by NADPH-dependent thioredoxin system. CSH, BioRixiv. doi.org/10.1101/261560. Sinclair et al. (2017) Etiolated Seedling Development Requires Repression of Photomorphogenesis by a Small Cell-Wall-Derived Dark Signal. Curr Biol. 2017 Nov 20;27(22):3403-3418.e7. doi: 10.1016/j.cub.2017.09.063. Gzyl et al. (2017). Gamma-tubulin distribution and ultrastructural changes in root cells of soybean (Glycine max L.) seedlings under cadmium stress. Environmental and Experimental Botany, Vol 143, Nov 2017, Pages 82-90. Kamies et al. (2017). A Proteomic Approach to Investigate the Drought Response in the Orphan Crop Eragrostis tef. Proteomes. 2017 Nov 15;5(4). pii: E32. doi: 10.3390/proteomes5040032. Niederhuber et al. (2017). Super-resolution microscopy of the ß-carboxysome reveals a homogenous matrix. Mol Biol Cell. 2017 Aug 9. pii: mbc.E17-01-0069. doi: 10.1091/mbc.E17-01-0069. Tyuereva et al. (2017). The absence of chlorophyll b affects lateral mobility of photosynthetic complexes and lipids in grana membranes of Arabidopsis and barley chlorina mutants. Photosynth Res. 2017 Apr 5. doi: 10.1007/s11120-017-0376-9. (Hordeum vulgare, western blot) Romanowska et al. (2017). Differences in photosynthetic responses of NADP-ME type C4 species to high light. Planta. 2017 Mar;245(3):641-657. doi: 10.1007/s00425-016-2632-1. Epub 2016 Dec 18. Good et al. (2016). Attenuating Listeria monocytogenes Virulence by Targeting the Regulatory Protein PrfA. Cell Chem Biol. 2016 Mar 17;23(3):404-14. doi: 10.1016/j.chembiol.2016.02.013. Datta et al. (2016). Glutathione Regulates 1-Aminocyclopropane-1-Carboxylate Synthase Transcription via WRKY33 and 1-Aminocyclopropane-1-Carboxylate Oxidase by Modulating Messenger RNA Stability to Induce Ethylene Synthesis during Stress. Plant Physiol. 2015 Dec;169(4):2963-81. doi: 10.1104/pp.15.01543. Epub 2015 Oct 13. Barahimipour et al. (2016). Efficient expression of nuclear transgenes in the green alga Chlamydomonas: synthesis of an HIV antigen and development of a new selectable marker. Plant Mol Biol. 2016 Mar;90(4-5):403-18. doi: 10.1007/s11103-015-0425-8. Epub 2016 Jan 8. CiteAB has listed this antibody as one of the 7000 most published antibodies in the world. |